Mickey Kosloff's lab at the Department of Human Biology, Faculty of Natural Sciences, The University of Haifa.
Our goals:
We focus on understanding how the �wiring together� of protein interaction networks drives cellular communication in health and disease. This is a fundamental challenge in studying and manipulating cellular signal transduction and in rational drug design � we would like to decipher how protein structure encodes interaction specificity, thereby determining how signaling proteins precisely recognize their interaction partners. To achieve these goals, our lab combines computational and experimental approaches in a multi-disciplinary strategy.
Our main research activities:
1) Understanding the molecular basis for protein-protein interaction specificity among large protein families.
2) Redesigning and engineering proteins as tools to perturb and modulate signaling networks in vivo.
3) Leveraging these insights and tools to address a critical need in drug design � pinpoint drug binding sites that take family-level specificity into account � thereby leading to more specific drugs with reduced side effects for diseases such as hypertension, arrhythmias, neurological disorders, and cancer.
To achieve these goals, we combine complementary computational and experimental approaches in a multi-disciplinary strategy. By combining the accuracy of experiments with the mechanistic insights and scalability of computations, we can extend our analysis to the level of whole families and eventually to rewiring of signaling networks at the cellular level.
Current Research projects:
1) Develop computational and experimental approaches to decipher the determinants of protein-protein interaction specificity at the family level.
2) Characterize the molecular basis for the function and modulation of molecular switches in G-protein coupled signaling networks by the RGS family.
3) Characterize the structural basis for specific protein-protein interactions between Receptor Tyrosine Kinases and their protein ligands.
4) Analyze the determinants of specificity in ligand binding sites.
5) Determine how interactions with membranes further modulate protein-protein interaction specificity.
6) Design custom-made signaling proteins to investigate and engineer how specificity wires signal transduction networks in vivo.
7) Structure-based drug design at the family-level and beyond.
8) Develop and apply a structure-based approach to search for pathogenic proteins that are novel and specific targets for next generation anti-pathogenic drugs.
More in-depth Background (not for the faint of heart):
The challenge of deciphering interaction specificity among signaling protein families:
For signaling cascades to function correctly, their components must be �wired� together accurately. This requirement presents a challenge for living cells, as similar components are used again and again in both parallel and intersecting cascades within the same cell. Signaling therefore requires that particular protein-protein interactions be tailored to each signaling cascade with either broad or narrow specificity. Understanding the basis for such selectivity is a major goal in biology, as well as in drug design. Yet, beyond single representative examples, little is known of how specificity is determined in the interactions between members of large protein families, including those involved in signal transduction.
Elucidating what determines the shared interactions or distinct specificities among large protein families is a formidable task. It is very common for a cell to contain many different members of large families that can co-localize and potentially interact with one another. Yet, while the number of potential combinations is very large in theory, because of interaction specificity the number of combinations that actually occur in vivo is surprisingly small. However, which interactions do occur and what determines the specificity of these interactions on the structural level is unclear. To complicate matters, considerable sequence diversity is common in many large protein families. Such low sequence identity increases the difficulty of pinpointing which residues determine similar interactions, and which residues contribute to selectivity.

Currently, computational methods are not up to the task of predicting either protein-protein or protein-membrane binding affinities, although such methods can provide residue-level structural insights and are easily scalable. On the other hand, while quantitative experimental comparisons offer superior accuracy, expanding such a comparative approach to the entire family level can be a daunting task, and will usually fail to yield mechanistic insights at the resolution of individual residues. Our solution to this conundrum is to combine the strengths of computations and experiments into a multidisciplinary approach.
RGS-G-proteins interactions - a model system to study interaction specificity:
The challenge of deciphering protein-protein interaction specificity is particularly relevant to the interactions of heterotrimeric (alpha-beta-gamma) G-proteins with Regulators of G-protein Signaling (RGSs). G-proteins are ubiquitous molecular switches that are essential for normal communication within and between cells (see the 1994 Nobel prize in Physiology or medicine and the 2012 Nobel prize in Chemistry). RGS proteins are the negative regulators of G-alpha subunits, functioning as GTPase Activating Proteins (GAPs). Namely, the RGS domain of ~120 amino acids, present in all RGS proteins, can bind specifically to certain GTP-associated G-alpha subunits and accelerate their intrinsic GTPase activity allosterically. Thereby, RGS proteins turn G-proteins �off� and determine the lifetime of the activated G-protein switch.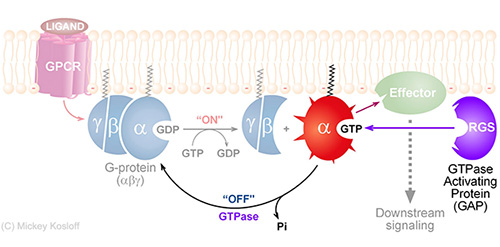
G-proteins and RGS proteins mediate countless cellular functions, including sensory transduction, hormonal action, proliferation and differentiation, cytoskeletal changes, and synaptic function. These large protein families have been implicated in a wide range of human pathologies, including hypertension, arrhythmias, schizophrenia, drug abuse, and cancer. Indeed, the G-protein and RGS families are promising drug targets, both as primary targets and as complements to drugs that target other components in G-protein signaling (such as G-protein Coupled Receptors). Many different members of the G-protein/RGS families are usually co-expressed in a given cell and are commonly co-localized at the cytosolic periphery of cellular membranes. In fact, often only a single family member (out of many co-expressed members) mediates a given biological function.
Importantly, which RGS�G-protein interactions do occur and which residues encode the specificity of these interactions is mostly unclear - partially due to the low sequence identity among human G-alpha subunits and RGS domains � 40% and 30%, respectively (indeed, high diversity is common to many signaling protein families). Such low sequence identity increases the difficulty of pinpointing which residues contribute to similar interactions, and which residues determine interaction selectivity. Therefore, identifying the specificity determinants of G-protein recognition by RGS proteins is essential for understanding these signaling pathways and for manipulating them with drugs.



